The Ogasawara Islands, also known as the Bonin Islands, are oceanic islands where evolutionary radiations of organisms, particularly in land snails and plants, have occurred (e.g., Ito 1998, Chiba 1999). The group consists of approximately 30 islands and forms part of the Izu-Bonin-Mariana arc system in the northwest Pacific Ocean. The Ogasawara Islands are located approximately 1,000 km south of Honshu Island, approximately 650 km south of the oceanic Izu Islands, approximately 750 km north of the Mariana Islands, and approximately 4,000 km west of the Hawaiian Islands. The land snail fauna of these islands is particularly diverse, with over 110 recorded indigenous species, exhibiting an exceptionally high level of endemism (more than 90%) (Chiba 2009a). The genus Hirasea Pilsbry, 1902 is endemic to the Ogasawara Islands and exhibits remarkable diversification in shell morphology among species (Chiba 2009b). This genus was originally described as a member of Zonitidae Mörch, 1864 within Zonitoidea Mörch, 1864, but has subsequently been assigned to multiple taxonomic groups. Some malacologists have classified Hirasea within Limacoidei, specifically under Trochomorphidae Möllendorff, 1890 and Euconulidae Baker, 1928 of Trochomorphoidea Mörch, 1864 (e.g., Kuroda 1930, Ueshima & Kurozumi 1988), as well as Helicarionidae Bourguignat, 1877 of Helicarionoidea Bourguignat, 1877 (Hirano et al. 2018). Others have placed it within Punctoidea Morse, 1864, assigning it to Endodontidae Pilsbry, 1895 or Charopidae Hutton, 1884 (e.g., Habe 1969, MolluscaBase 2021). Still others have classified it within Discidae Thiele, 1931 of Discoidea Thiele, 1931 (e.g., Kuroda 1958). To date, molecular phylogenetic approaches have been applied to this genus in two studies. Hirano et al. (2018) focused on the phylogenetic relationships within Hirasea and used taxa classified within Helicarionidae as an outgroup. Colgan & Stanisic (2023) primarily included Punctoidea in their analysis and failed to identify any taxon closely related with Hirasea. Therefore, the taxonomic position of Hirasea remains unresolved. In this study, we conducted a phylogenetic analysis of the genus using a broad range of stylommatophoran land snail species to clarify its taxonomic position.
Specimens of Hirasea (Figs 1–2) were collected on the Ogasawara Islands under permits issued by the Agency for Cultural Affairs and the South Kanto branch, Ministry of the Environment. Additionally, specimens from ten genera belonging to Trochomorphoidea were collected from the Japanese mainland (Table 1). The specimens examined in this study were identified based on the taxonomic descriptions provided in the literature including Azuma (1982), Iijima (2018) and Nishi & Nishi (2018). The specimens used in this study were preserved in 99.5% ethanol and deposited in the collection of Tohoku University (Table 1). Genomic DNA was isolated from a piece of foot using DNeasy Blood and Tissue kits (Qiagen, CA, USA) according to the manufacturer’s instructions. Partial fragments (about 1,000 bp) of the large subunit 28S rRNA gene were amplified and sequenced using the universal primers 28SC1 and 28SD3 (Vonnemann et al. 2005). The condition used for the polymerase chain reaction (PCR) was as follows: 94 °C for 3 min, (94 °C for 30 s, 50 °C for 30 s and 72 °C for 1.25 min) × 35, and 72 °C for 5 min. The PCR products were purified using Exo-SAP-IT (Amersham Biosciences, Little Chalfont, Buckinghamshire, UK). Sequencing was performed using an ABI 3130xl sequencer (Applied Biosystems, Carlsbad, CA, USA). The resulting 28S sequences have been deposited in the DDBJ/EMBL/GenBank database (Table 1). In addition to these newly obtained sequences, sequence data from Wade et al. (2001), which analysed a wide range of stylommatophoran species, were obtained from GenBank and included in our phylogenetic analyses (Appendix 1).
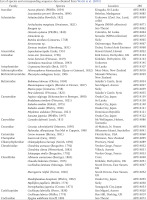
Figs 1–2
Living animal and shell morphology of Hirasea: 1 – Hirasea operculina; 2 – Hirasea diplomphalus (photographed on 14 March 2009 at Anijima Island, Tokyo, Japan)

DNA sequences were aligned with MUSCLE v3.8 (Edgar 2004). To eliminate any uncertainty in the 28S, trimAl 1.2 with automated option (Capella-Gutiérrez et al. 2009) was used to exclude ambiguous alignment regions. Phylogenetic trees were constructed for the 28S dataset (375 bp) using maximum likelihood (ML) and Bayesian inference (BI) methods. Prior to the ML and BI analyses, ModelFinder (Kalyaanamoorthy et al. 2017) was used to select the appropriate models for sequence evolution. As a result, the following models were selected: GTR+F+R4 in the ML analysis; GTR+G in the BI analysis. ML analysis was performed with IQ-TREE (v. 1.6.12) using the option of a perturbation strength of 0.7 (Nguyen et al. 2015, Chernomor et al. 2016). For the ML analyses, we assessed nodal support by performing ultrafast bootstrap analyses with 10,000 replications (Hoang et al. 2018). BI analysis was performed using MrBayes (v. 3.1.2) with two simultaneous runs (Ronquist & Huelsenbeck 2003). Each run consisted of four simultaneous chains for 20 million generations and sampling of trees every 1,000 generations. We confirmed the convergence of runs and sufficient effective sample sizes using Tracer (v. 1.6; Rambaut et al. 2013). Then we discarded the first 2,000 trees as burn-in and used the remaining samples to estimate tree topology, branch length, and substitution parameters.
The phylogenetic analyses using ML and BI methods yielded phylogenetic trees with nearly identical topology. Only well-supported clades (ultrafast bootstrap support values ≥ 95% or posterior probabilities ≥ 0.95) are considered hereafter. Our phylogenetic analyses showed that species of Hirasea belong to Limacoidei, rather than to Punctoidea or Discoidea (Fig. 3). Hirasea was placed in Clade A together with Euconulidae and Chronidae Thiele, 1931, whereas Trochomorphidae was not included in this clade (Fig. 3). However, our analyses did not support the monophyly of either Euconulidae or Chronidae. This result is consistent with the findings of Pholyotha et al. (2023), who focused on Euconulidae in Thailand. Therefore, a taxonomic revision of both Euconulidae and Chronidae is needed to determine the family to which Hirasea belongs. Our analyses included only a few species from Punctoidea and Discoidea, and did not include Geotrochidae Schileyko, 2002 and Staffordiidae Thiele, 1931 of Trochomorphoidea. Since Staffordiidae is restricted to the Dafla Hills at the foot of the Eastern Himalayas and remains a poorly understood family (Hausdorf 2000), we are unable to discuss this family in detail in this study. The monophyly of each of Punctoidea, Discoidea, and the individual families within Trochomorphoidea – except for Euconulidae, Chronidae, and Staffordiidae – has been strongly supported by molecular analyses (Salvador et al. 2020, Pholyotha et al. 2023). Therefore, the inclusion of additional species from these taxa is unlikely to affect the conclusions drawn in the present study.
Fig. 3
Maximum likelihood tree of stylommatophoran land snails based on the 28S rRNA gene. Asterisks on branches indicate high ultrafast bootstrap values (≥ 95%, left) and Bayesian posterior probabilities (≥ 0.95, right). The shaded box highlights species of Hirasea. Filled triangles, filled squares, and open square represent Euconulidae, Chronidae, and Trochomorphidae, respectively. Clade A represents a well-supported clade comprising Hirasea, Euconulidae, and Chronidae
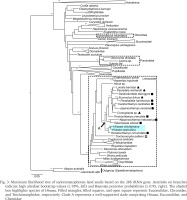
Our analyses also revealed a taxon closely related with Hirasea on the Japanese mainland. The ancestors of certain indigenous land snails of the Ogasawara Islands are thought to have been derived from the Japanese mainland (e.g., Chiba 1999), and this could be the case for the focal genus. However, dispersal from the Oceanian Islands has also contributed to the development of the land snail fauna on the Ogasawara Islands (e.g., Goulding et al. 2023). Therefore, further analyses using additional specimens from both the Japanese mainland and the Oceanian Islands are needed to infer the route by which the ancestors of Hirasea reached the Ogasawara Islands.
This study provided preliminary insights into the molecular phylogenetic relationships of Trochomorphoidea in Japan, although approximately half of Japanese genera of this superfamily remain to be examined in future studies. For example, Horsáková et al. (2020) suggested that the Holarctic genus Euconulus Reinhardt, 1883 could have a close relationship with Parakaliella Habe, 1946 based on similarities in shell morphology, and this hypothesis was supported by our findings (Fig. 3).