INTRODUCTION
The family Cochliopidae Tryon, 1866 constitute a group of cosmopolitan freshwater snails, which are usually of a small size. It comprises ca. 70 genera and 3 subfamilies (MolluscaBase 2022) of which some members of the subfamily Semisalsinae Giusti et Pezzoli, 1980 are distributed in Europe. Members of this subfamily mainly inhabit springs, streams and some river sections, all with permanent and fast-flowing water, and they are often associated with a high degree of salinity (Schütt 1991, De Francesco & Isla 2003, Martin 2008).
The subfamily Semisalsinae had as its type genus Semisalsa Radoman, 1974, which was proposed for the European species of the genus Heleobia Stimpson, 1865. However, an older name was erected before, Eupaludestrina Mabille, 1877, making Semisalsa a junior synonym (Kadolsky 2012) thus being the accepted name Eupaludestrina according to the principle of priority (Alvarado 1962).
The genus Eupaludestrina currently comprises 19 extant species (MolluscaBase 2022), which were mainly described in the century 19th and early 20th century, sometimes with a very short description, imprecise type localities and no illustrations. That is the case for Eupaludestrina canariensis (Mousson, 1872) described from the vicinity of “Ríos Palmas”, Fuerteventura, Canary Islands, Spain. Originally treated as a member of the genus Hydrobia W. Hartmann, 1821, the species was erected after the analysis of only two shells (Mousson 1872) and there is no plate or image corresponding to either of these individuals in the subsequent publication. The species has remained poorly known, with only a few mentions of its existence in international databases and the Canary Island archipelago fauna checklist (Arechavaleta et al. 2010, MolluscaBase 2022, WoRMS 2023).
The present work aims to redescribe the species Eupaludestrina canariensis and analyse the genetic variability of the genus including all sequenced European congeners, and at the same time place Eupaludestrina canariensis in a phylogenetic context to use an integrative taxonomy approach.
MATERIALS AND METHODS
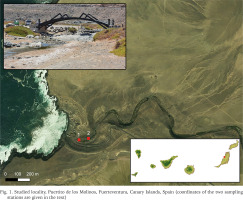
One population of Eupaludestrina has been found in Puertito de los Molinos, Fuerteventura, Canary Islands, Spain (Fig. 1), which, given its geographical proximity to the locus typicus, can reasonably be regarded as Eupaludestrina canariensis. The specimens were collected on June 2021 in a stream that flows directly to the ocean. Two different sampling stations were selected where the animals were found living in shallow waters, following the stream course upwards (1: 28°32'33.0″N, 14°03'46.2″W and 2: 28°32'32.9″N, 14°03'42.6″W). After collection, specimens were transferred to 96% ethanol. Once in the laboratory, pictures were taken using a Leica MZ16 stereomicroscope with a Leica DFC550 camera using the Leica Application Suite (LAS) V4.6.
Fig. 1
Studied locality, Puertito de los Molinos, Fuerteventura, Canary Islands, Spain (coordinates of the two sampling stations are given in the text)
Due to the small size of the specimens, DNA was extracted using the whole animal. Isolation was conducted using a DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany). For DNA amplification the mitochondrial cytochrome c oxidase subunit I (COI) was used with the following primers: LCO1490 (Folmer et al. 1994) and COR722b (Wilke & Davis 2000). PCR was realised under the following conditions: each tube contained 1 µl DNA, 2 µl of 10× Buffer, 0.8 µl dNTPs mix (each base with the same concentration), 0.25 µl MgCl2 25 mM solution, 0.5 of each primer (10 mM), 0.3 µl Taq DNS polymerase (5 U/µl – Takara) and 19.65 µl purified distilled water to complete 25 µl of mix for reaction. The cycling conditions were 94 °C for 4 min, one cycle; 94 °C for 45 secs, 48 °C for 45 secs, 72 °C for 45 secs, 35 cycles; 72 °C for 10 min for the final extension, after that kept at 4 °C. One microliter of the PCR product was analysed to determine the quantity of DNA obtained from the PCR. It was verified using 1% agarose gel with SYBR Safe (Invitrogen, USA) to visualise it under UV light. After that 10 µl of the PCR product was cleaned using EXOSAP (ThermoFisher, USA), and then 5 µl of PCR product plus primer (5 mM – Forward and Reverse) were sequenced at Macrogen (Macrogen, Korea) based in Madrid.
The obtained sequences were edited using Sequencher v.5.4.6 (Gene Codes, Ann Arbor, MI, USA). Other available sequences from GenBank (Clark et al. 2016) were downloaded using R 3.5.3 (R Core-Team 2022) with the ape (Paradis & Schliep 2018), seqinr (Charif & Lobry 2007) and rentrez (Winter 2017) packages (see Table 1 for more details). Among all the sequences of the family Cochliopidae available on GenBank, a first selection was done according the taxonomic criterion, including as much as possible the best representation of the family. Posteriorly the initial matrix was trimmed, removing those sequences that presented incongruences with the taxonomic position assigned by GenBank (i.e., sequences assigned to one genus that grouped with different genera in the initial phylogenetic analyses) leaving only the sequences relevant for this work and those without taxonomical errors that could lead to erroneous conclusions.
Table 1
Species, Locality and GenBank Accession number of all the sequences used for the molecular studies

As COI is a protein-coding gene fragment, all the sequences were in the next step unambiguously aligned manually using MEGA v.7.0.14 (Kumar et al. 2016). The species Trichonia kephalovrissonia Radoman, 1973 and Beddomeia krybetes Ponder et Clark, 1993 were used as outgroups.
In order to determine the level of substitution saturation on the COI dataset a saturation test (Xia et al. 2003, Xia & Lemey 2009) was implemented in DAMBE v. 7 (Xia 2018). Significant values of this test (p < 0.001) indicate little saturation; thus, the data contain phylogenetic information. Sequence divergences (uncorrected p-distances) were calculated in MEGA v.7.0.14.
Phylogenetic relationships of Eupaludestrina were assessed for the COI dataset under Bayesian inference (BI) and maximum likelihood (ML). The BI analysis was conducted using Markov chain Monte Carlo (MCMC) sampling in MrBayes v.3.2.7a (Ronquist et al. 2012) under the mixed substitution models. The MCMC sampling was run for 20 × 106 generations, four parallel chains and saving a tree every 1,000 generations. After assessing chain convergence by ensuring a standard deviation of the split frequency < 0.01, the first 10% of the sampled trees were discarded as burn-in. The robustness of the inferred tree was quantified using Bayesian posterior probabilities (BPP). The ML analyses were computed with RAxML-NG v.1.0.2 (Kozlov et al. 2019), with 10 random starting trees, applying the best-fit substitution model, selected by jModelTest v.2.1.6 (Darriba et al. 2012) according to the corrected Akaike information criterion (AIC) (Akaike 1974). The selected model was: HKY (Hasegawa et al. 1985)+ I + Γ.
Branch supports were assessed by heuristic bootstrapping (BS) with a stopping threshold of 0.03 and later quantified using the transfer bootstrap expectation (TBE; Lemoine et al. 2018) metric. Final trees and branch supports were visualised in FigTree v.1.4.3 (Rambaut 2012).
To better represent genealogies at smaller scales, a haplotype network was calculated using the unrooted COI matrix selecting only Eupaludestrina species. A TCS network (Clement et al. 2000) was constructed in PopART software (Population Analysis with Reticulate Trees; Leigh & Bryant 2015) setting all parameters as default and a different colour for each of the biogeographical region. The biogeographical regions were coded as a trait in PopART according to the Freshwater Ecoregions of the world (Abell et al. 2008). The current distributions of the species were obtained from international fauna databases and the original descriptions of the species (De Stefani 1883, Radoman 1974, Boeters et al. 1977, MolluscaBase 2022, Worms 2023).
RESULTS
PHYLOGENETIC RELATIONSHIP OF EUPALUDESTRINA SPP.
Analysis of the mitochondrial COI dataset yielded an alignment of 655 bp covering a total of 32 specimens of Cochliopidae. Base frequency was T 39.1%, C 17.5%, A 23.6% and G 19.8%. From the complete dataset, 59.6% were invariant sites.
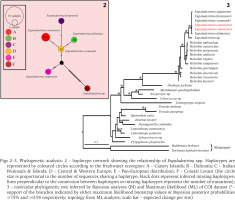
In both ML and BI, Eupaludestrina resulted in a monophyletic group, with Heleobia as a sister group. Mean sequence divergence between Eupaludestrina species ranged from 0.31% between E. scamadri and E. foxianensis to 6.94% between E. stagnorum and Eupaludestrina sp. (Table 2). The COI sequences of the whole dataset grouped in 6 haplotypes according to the TCS network, recovering E. canariensis (Fig. 2 H4) as a single haplotype.
Table 2
Genetic distance matrix (uncorrected p-values) based on a fragment of the COI gene (655 bp.) expressed in per cent
Figs 2–3
Phylogenetic analysis: 2 – haplotype network showing the relationship of Eupaludestrina spp. Haplotypes are represented by coloured circles according to the freshwater ecoregion: A – Canary Islands; B – Dalmatia; C – Italian Peninsula & Islands; D – Central & Western Europe; E – Pan-European distribution; F – Coastal Levant (the circle size is proportional to the number of sequences sharing a haplotype; black dots represent inferred missing haplotypes; lines perpendicular to the connection between haplotypes or missing haplotypes represent the number of mutations); 3 – molecular phylogenetic tree inferred by Bayesian analyses (BI) and Maximum likelihood (ML) of COI dataset (*– support of the branches indicated by either maximum likelihood bootstrap values or Bayesian posterior probabilities >75% and >0.95 respectively; topology from ML analysis; scale bar – expected change per site)
SYSTEMATICS
Class GastropodaCuvier, 1795
Subclass CaenogastropodaCox, 1960
Order Littorinimorpha Golikov et Starobogatov, 1975
Superfamily TruncatelloideaGray, 1840
Family CochliopidaeTryon, 1866
Subfamily Semisalsinae Giusti et Pezzoli, 1980
Genus EupaludestrinaMabille, 1877
Eupaludestrina canariensis (Mousson, 1872)
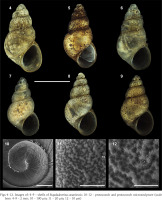
Figs 4–12
Images of: 4–9 – shells of Eupaludestrina canariensis; 10–12 – protoconch and protoconch microsculpture (scale bars: 4–9 – 2 mm; 10 – 100 µm; 11 – 20 µm; 12 – 10 µm)
Hydrobia canariensis Mousson, 1872: pp. 148–149.
Heleobia canariensis (Mousson, 1872)
Diagnosis. Shell conical, elongated. Protoconch microsculpture pitted; teleoconch whorls convex, separated by a deep suture; body whorl large, convex, occupying more than two-thirds of the total shell length; rachidian tooth with only 1 basal cusp. Female genitalia with a capsule gland divided into two pigmented regions, the distal part yellowish and the proximal part with a terracotta colour characteristic of the genus.
Examined material. Specimens from Puertito de los Molinos, Fuerteventura, Canary Islands, Spain.
Type locality. “Dans une mare près du Rios Palmas, Fuerteventura”. In a pool near Río Palmas, Fuerteventura, Canary Islands, Spain, according to the original description.
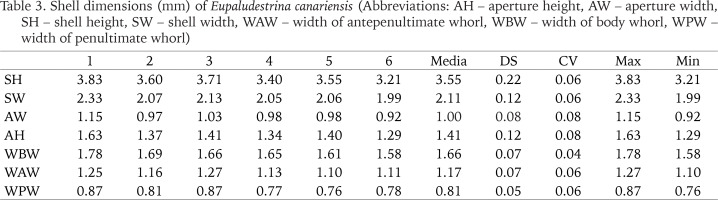
Description. Shell conical, elongated with moderately pointed apex; whorls 4–5, height 3.5 ± 0.2, (range 3.21–3.83) mm, width 2.01–2.36 mm (Figs 4–9; Table 3); brown periostracum; protoconch of 1.5 whorls, ca. 270 µm wide, nucleus ca. 190 µm wide; protoconch microsculpture pitted (Figs 4–6); teleoconch whorls convex, separated by a deep suture; body whorl large, convex, occupying more than two-thirds of the total shell height; aperture obliquely broad ovate, complete; inner lip thicker than outer lip; aperture margin straight, inner lip touching the shell wall; umbilicus narrow, covered by the inner lip. Operculum yellowish, about two whorls; muscle attachment oval, located near the nucleus.
Table 3
Shell dimensions (mm) of Eupaludestrina canariensis (Abbreviations: AH – aperture height, AW – aperture width, SH – shell height, SW – shell width, WAW – width of antepenultimate whorl, WBW – width of body whorl, WPW – width of penultimate whorl)
Radula central tooth formula 5(6)–C–5(6)/1–1, central cusp “V” shaped, cutting edge slightly concave (Figs 21–22). Lateral tooth formula 5(4)–C–5(4), central cusp “V” shaped and slightly longer than the central tooth one. Inner marginal teeth with 19–24 cusps; outer marginal teeth with more than 24 cusps.
Figs 13–22
Anatomy of Eupaludestrina canariensis, specimens from Puertito de los Molinos, Fuerteventura, Spain: 13 – animal depicting body pigmentation; 14 – ctenidium and osphradium; 15 – stomach; 16 – perioesophageal ring; 17–20 – female reproductive system; 21 – fragment of the radular ribbon, showing rachidian tooth, lateral and marginal row of teeth; 22 – detailed view of the lateral and marginal teeth. Abbreviations: Ag – albumen gland, Bc – bursa copulatrix, Cg – capsule gland, Ct – ctenidium, Os – osphradium, Ro – renal oviduct, Sr1 – seminal receptacle. Scale bars: 13, 14, 17 – 1 mm; 15, 16, 19, 20 – 500 µm; 21 – 50 µm; 22 – 20 µm
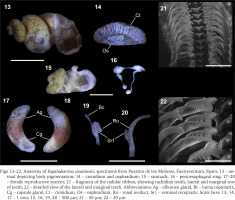
Animal darkly pigmented (Fig. 13); head and tentacles black, with a white line from the base to the tip of the tentacles; eye lobes and neck, less pigmented; snout about as long as wide, approximately parallel-sided, with medium distal lobation (Fig. 13). Ctenidium occupying almost the total length of the pallial cavity; 21–27 gill filaments; filaments broad, triangular, fused at the base by a vessel. Osphradium elongate, more than 3 times longer than wide, positioned opposite to the middle of the ctenidium (Fig. 14). Stomach with two chambers almost equal in size; style sac with the unpigmented intestine surrounding its distal end before continuing as a straight rectum (Fig. 15).
Female genitalia with a glandular oviduct covered by a pigmented epithelium; glandular oviduct 2.5 times longer than wide; albumen gland shorter than capsule gland; capsule gland divided into two pigmented regions the distal part yellowish and the proximal part with a terracotta colour characteristic of the genus; bursa copulatrix minute, pedunculated with a long duct, renal oviduct unpigmented; seminal receptacle elongate, with a short duct, positioned at the distal end of the renal oviduct just above the junction with the bursal duct (Figs 19–20). Nervous system unpigmented (Fig. 16), elongated (mean RPG ratio – 0.66); cerebral ganglia approximately equal in size; pleuro-supraoesophageal connective ca. 8 times longer than the pleuro-suboesophageal one.
Remarks. The author of the species relates that his first impression was that the two specimens analysed corresponded to Hydrobia ventrosa, but the values of the length and width that he gives are far from the values that Montagu (1803) gives for this species (height – 3.17 mm; width – 1.04 mm, very imprecise considering that the original values are expressed in inches). Abnormally large specimens are commonly found among hydrobioids (i.e., informal group to refer to species similar in shell characteristics to members of Hydrobiidae) and it can depend on a variety of factors such as flow rate and turbidity of the stream (Verhaegen et al. 2018) or the relative age of the animal, generally univoltine and semelparous (i.e., animals with just one generation per year and that breed once in a lifetime). The values found in the analysed samples are closer to the genus Eupaludestrina according to Radoman (1974) (Table 1).
The radula presented one suitable character to discriminate among species, as E. canariensis presented just one pair of basal cusps in the rachidian tooth (Fig. 21) and according to Radoman (1974) the European species (i.e., E. dalmatica Radoman, 1974 and E. rausiana (Radoman, 1974)) presented 2–4 pairs of basal cusps in the rachidian tooth.
Mousson (1872) did not illustrate his species. A supposed illustration of the species, given in WoRMS (2023) is of a specimen collected in Caleta del Marrajo, Fuerteventura, Canary Islands, Spain. This locality is marine, and the specimen illustrated corresponds more to a marine rissooidean than to a brackish truncatelloidean.
The type locality is located in one of the few permanent freshwater streams on the island. In the surveys performed for this work, the final part of the stream Rio Palmas was found dry as a consequence of the dams in the upper watercourse. In addition, the species was not found in the pool near Vega de Río Palma, Fuerteventura, Spain.
Ecology and Distribution. The species occurs in brackish environments as the other members of the genus, only known at the type locality and now in Puertito de los Molinos, Fuerteventura, Spain.
DISCUSSION
Originally Cochliopidae was treated as a member of Hydrobiidae s.l., being upgraded to the family level after the work of Wilke et al. (2013). We lack a complete phylogeny of the family, although the work of Liu et al. (2001) established the monophyly of Cochliopinae (within Hydrobiidae). Molecular phylogenies are found more often in works on the species occurring in the Americas (e.g., Alvear et al. 2020, Collado & Fuentealba 2020, Collado et al. 2020) rather than in species occurring in Europe. However, the results here presented recovered Eupaludestrina as the sister group of Heleobia as in Collado et al. (2020).
Phylogenetically, the species of Eupaludestrina show low COI divergence distances (uncorrected p-distances < 2%).While divergences above 5% are often used to distinguish good species, there are cases were interspecific COI divergence can be far below 5% (Haase et al. 2007). Rissooidean and truncatelloidean gastropods are characterised by its little genetic variation in general, correlated with some degree of morphological differentiation. For instance, a threshold of 1.3% has been observed between species of Mercuria Boeters, 1971 (Miller et al. 2022), 3% was found in Hydrobia (Wilke et al. 2000), 1.5% in Bythinella Moquin-Tandon, 1856 (Bichain et al. 2007) and 0.5% in Floridobia Thompson et Hershler, 2002 (Hershler et al. 2003). The COI threshold could be regarded as an efficient tool to assign diversity; however, it must be considered cautiously since it needs to be confronted with additional data (Bichain et al. 2007), in our case morphological data was used.
The haplotype network also supports this low variability in COI, finding a small number of mutations between haplotypes H1–H4. Similar low divergence had been found in Bythinella as the result of allopatric speciation during glacial stages (Benke et al. 2009). It is probably the same process for the European species of Eupaludestrina since all freshwater environment were subjected to the same conditions during glacial periods.
All the European species of Eupaludestrina are rather similar in shell size and shape, and in general, freshwater hydrobioids present low variability in shell shape (Wilke et al. 2000, Haase 2005). However, when analysing the radular teeth, a suitable diagnostic character for E. canariensis seems to be the number of basal cusps in the rachidian tooth, which distinguishes it from other European species.
Since there are neither specimens nor illustrations for Mousson’s species, the validity of the taxon rests on the lack of any other permanent freshwater course on Fuerteventura where such species can occur, since the type locality has been rendered sterile. The radular difference distinguishes it from others.
Further studies including all the European species of Eupaludestrina are needed to clarify the status of currently recognised species; in effect, to determine whether the known morphological variation is merely intraspecific. The results presented here form the basis for a more comprehensive, integrative revision, that might reduce many species into synonymy.

