INTRODUCTION
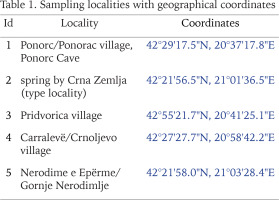
Radoman (1978) described a new genus of the truncatelloid snail Terranigra Radoman, 1978, with its type species Terranigra kosovica Radoman, 1978. The genus still remains monotypic (Radoman 1983, 1985, Kabat & Hershler 1993). The genus was characterised by a big bursa copulatrix with a short duct, two seminal receptacles, with the distal (rs1) seminal receptacle very small, nearly vestigial, and the proximal (rs2) seminal receptacle big, and the penis simple without outgrowths. The type locality of T. kosovica is a spring by Crna Zemlja (which means black soil, in Latin terranigra), near Mala Reka, about 6 km west of Nerodimlje (=Nerodime e Epërme) and 12–13 km west of Uroševac (=Ferizaj) town (Table 1, Fig. 1, locality 2). The species was reported by Radoman (1983) from three other additional localities (Table 1, Fig. 1): a small spring at the village of Pridvorica, near the place Velji Breg on the road Ribarić-Kosovska Mitrovica (locality 3); the spring below the rock in Crnoljevo village (=Carralevë), Gornja Mahala, a little over the road Štimlje-Prizren (locality 4); the spring (fountain) at the forester’s hut, near the confluence of the Mala and Velika Reka, at the beginning of Nerodimka River (locality 5) (Radoman 1978). This species, with an unpigmented body and with eyespots present, was reported as a “subterranean form” inhabiting also epigean springs (Radoman 1978, 1983). To our knowledge, there are no more published data on Terranigra, except imprecise information about 10 sites in The IUCN Red List of Threatened Species (Seddon 2011).
Fig. 1
Geographic distribution of the known localities of Terranigra kosovica. Inset indicates the location of the study area. For country identification we used the officially assigned code ISO (ISO 3166 alpha-2)
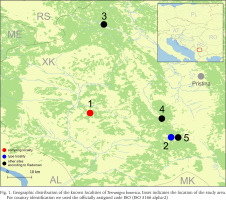
In the autumn of 2018, a new attempt was made to find this species in some of its known localities. As in previous years, we were unsuccessful in our attempt to find the locus typicus as described by Radoman. We had equally negative results in two other localities listed by Radoman (1983), but we did discover a previously unknown Terranigra population near Ponorc/Ponorac village.
The aim of the present study was to collect material for genetic analysis to resolve issues with the systematics of these snails. The availability of living specimens has allowed us to use DNA-based methods to identify the position of the species (and the monotypic genus) in the phylogenetic tree.
MATERIAL AND METHODS
Our field activities in Kosovo in years 2015–2018 includes a freshwater gastropod diversity survey over 96 springs of the region, many of them repeatedly sampled. During the end of September 2018 field trip, we sampled springs at 20 different locations. Two of which were thought to correspond to localities known from the literature for Terranigra kosovica (Table 1, Fig. 1). One was the locality west of Nerodime e Epërme/Gornje Nerodimlje, at beginning of the Nerodime/Nerodimka River (some springs along a ca. 500 m section downstream from the confluence with Mala Reka), 690 m a.s.l., 42°21'58.0″N, 21°03'28.4″E (locality 5). The other one was at Carralevë/Crnoljevo village from the large karstic spring near the mosque, 640 m a.s.l., 42°27'27.7″N, 20°58'42.2″E (locality 4). Despite careful searching (including digging and filtering in spring zone, picking underside of the stones and fine sandy sediment sieving), T. kosovica was not found at these historical localities. This of course does not provide conclusive evidence, but strongly suggests that the species has become extirpated at these localities since 1978. At one of the other 18 sampled springs at this field trip we found a population that we could identify as T. kosovica. This locality was south of Ponorc/Ponorac village (Prizren District), in side-springs beneath Ponorc Cave, 575 m a.s.l., 42°29'17.5″N, 20°37'17.8″E (Table 1, Figs 1–3, locality 1). Some animals were hand collected from the bottom side of larger stones lying in the spring, some others were found in by sieving the sandy sediment of the spring head as described in Grego et al. (2017a). Sediment samples were sieved through a 3 mm and 500 μm sieve and fixed in 80% analytically pure ethanol, replaced two times, and later the samples were sorted. Next, the snails were put in fresh 80% analytically pure ethanol and kept in −20 °C temperature in a freezer. The shells were photographed with a CANON EOS 50D digital camera, under a NIKON SMZ18 microscope with dark field.
Figs 2–3
Ponorc Cave: 1 – distant view of the hillside where Ponorc Cave is located, 2 – one of the small springs beneath Ponorc Cave (locality 1)

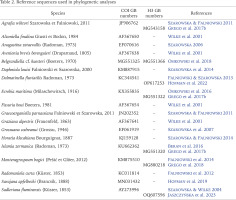
DNA was extracted from whole two specimens; tissues were hydrated in tris-EDTA (TE) buffer (3 × 10 min); then total genomic DNA was extracted with the SHERLOCK extraction kit (A&A Biotechnology), and the final product was dissolved in 20 μl of TE buffer. The extracted DNA was stored at −80 °C at the Department of Malacology, Institute of Zoology and Biomedical Research, Jagiellonian University in Kraków (Poland). Fragments of the mitochondrial cytochrome oxidase subunit I (COI) and the nuclear histone H3 (H3) were sequenced. Details of PCR conditions, primers used, and sequencing technique were given in Szarowska et al. (2016). The Sanger sequencing, in both directions, was performed at Genomed Company in Warsaw, Poland.
In the phylogenetic analysis additional, most similar sequences from GenBank were used (Table 2). Sequences of both loci were initially aligned with MUSCLE (Edgar 2004) program implemented in MEGA 7 (Kumar et al. 2016). The correctness of the alignment was checked in BIOEDIT 7.2.5 (Hall 1999), this program was also used to check for reading frame and stop codons. Uncorrected mean p-distances were calculated in MEGA 7. We analysed COI and H3 datasets individually, as well as concatenated dataset (COI+H3). For all datasets, the estimation of the proportion of invariant localities and the saturation test (Xia 2000, Xia et al. 2003) were performed using DAMBE (Xia 2018). The phylogenetic analysis was performed applying two approaches: Bayesian Inference (BI) and Maximum Likelihood (ML). In the BI analysis, the GTR+I+Γ model of nucleotide substitution was applied. Model was selected using MRMODELTEST 2.3 (Nylander 2004). The Bayesian analyses were run using MrBayes v. 3.2.3 (Ronquist et al. 2012) with defaults priors. Two simultaneous analyses were performed, each with 10,000,000 generations, with one cold chain and three heated chains, starting from random trees and sampling the trees every 1,000 generations. The first 25% of the trees were discarded as burn-in. The analyses were summarised as a 50% majority-rule tree. Convergence was checked in Tracer v. 1.5 (Rambaut & Drummond 2009); in all cases the effective sample size exceeded 200. FigTree v.1.4.4 (Rambaut 2010) was used to visualise the trees. The maximum likelihood analysis was conducted in RA×ML v. 8.2.12 (Stamatakis 2014) using the ‘RA×ML-HPC v.8 on XSEDE (8.2.12) tool via the CIPRES Science Gateway (Miller et al. 2010). We used the GTR+G model, using jModelTest2 via CIPRES.
RESULTS
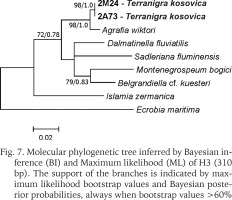
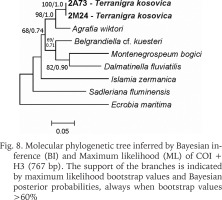
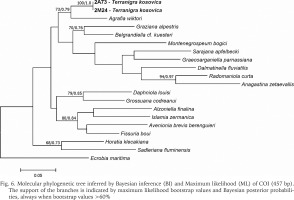
The examined animals and their anatomy were nearly identical with the one described by Radoman (1978); the unpigmented body (Figs 4–5), the soft part morphology and anatomy confirmed identification as T. kosovica. We obtained two new sequences of COI (457 bp, GenBank accession numbers OR373065-OR373066) and two of H3 (310 bp, GenBank accession numbers OR471608-OR471609). The tests by Xia et al. (2003) revealed no saturation. Results from the substitution saturation analysis showed an ISS (0.76 for COI; 0.50 for H3) substantially smaller than the critical ISS value (ISSC: 0.96 for COI; 0.62 for H3), indicating that all sequences are useful in phylogenetic reconstruction. The topologies of the trees inferred with ML and BI approaches were identical (Figs 6–8). At each tree (for COI, H3 and concatenated sequences) the sister genus of Terranigra was Agrafia Szarowska et Falniowski, 2011 (p-distance for COI was 0.074 and for H3 was 0.010). The bootstrap support and Bayesian posterior probability for concatenated sequences were high (98%, and 1.0 respectively). Islamia Radoman, 1973 was placed far from these two sister species in each tree, although in the COI tree (Fig. 6), as expected, basal nodes are not supported. In the H3 tree (Fig. 7), however, Islamia was outside of the well supported (bootstrap 72%) clade including Terranigra, Agrafia, Dalmatinella Radoman, 1973, Sadleriana Clessin, 1890, Montenegrospeum Pešić et Glöer, 2013, and Belgrandiella A. J. Wagner, 1928. Nearly the same relationships are present in the tree based on the concatenated sequence (Fig. 8), only the bootstrap is lower (68%), and Sadleriana does not belong to this clade.
Fig. 6
Molecular phylogenetic tree inferred by Bayesian inference (BI) and Maximum likelihood (ML) of COI (457 bp). The support of the branches is indicated by maximum likelihood bootstrap values and Bayesian posterior probabilities, always when bootstrap values >60%
DISCUSSION
As stressed in many publications, in Truncatelloidea, morphological data are sufficient neither for species distinction (Falniowski 2018, Chertoprud et al. 2023), nor higher classification (e.g. Szarowska & Falniowski 2008). Despite several efforts to apply molecular data (e.g. Wilke et al. 2001, 2013) we are still far from a well-supported phylogeny of the Truncatelloidea, including the Hydrobiidae. Our study confirmed the genus level status of Terranigra: genetic divergence between this species and its sister genus was 7.4%. This value is higher than interspecies level in many Hydrobiids (e.g. 1.5 to 3.5% estimate for interspecies divergence for Bythinella: Bichain et al. 2007). In our trees Terranigra clearly belongs to the family Hydrobiidae Stimpson, 1865. The well supported clade based on H3 represents the subfamily Sadlerianinae sensu Szarowska, 2006 (not Sadlerianinae Radoman, 1973). This name was proposed by Szarowska (2006) as a provisional replacement name for Horatiinae D. W. Taylor, 1966, since the specimen whose sequence was the only one available at that time in GenBank for “Horatia klecakiana” was the one used in the phylogeny of the Rissooidea by Wilke et al. (2001), and this specimen was misidentified (Szarowska & Falniowski 2014). It has to be pointed out that Sadlerianinae sensu Szarowska, 2006 was used for the clade sister to the Hydrobiinae (the latter including also Pyrgula and Pseudamnicola), grouping most of the genera of the Hydrobiidae. The more recent systematics of Wilke et al. (2013) followed by Bouchet et al. (2017) lists several subfamilies within the Horatiinae sensu Szarowska, 2006, Belgrandiellinae Radoman, 1973, Horatiinae D. W. Taylor, 1966 and Islamiinae Radoman, 1973 among them, but not resolving the relationships between them, showing either polytomy or paraphyletic Islamiinae. Clearly, our phylogeny based on COI and H3, is not congruent with the one presented in WoRMS (2022), which without any justification classify Agrafia within the subfamily Islaminae Radoman, 1973, and Terranigra within the subfamily Belgrandiinae de Stefani, 1877, thus sister taxa in our tree classifying in two subfamilies, confirming that a credible phylogeny of the Truncatelloidea is badly needed.
The last published IUCN Red List of Threatened Species (Seddon 2011) ranked this species as Near Threatened (NT). At that time, the number of locations was thought to be 10 and the Extent of Occurrence (EOO) were estimated as the whole area of Kosovo. The fact that there is good reason to believe that some populations have been extirpated since 1978 and that the main threats to the species’ habitats, namely habitat conversion and water abstraction, will continue in the future, would qualify this species as Endangered (EN) based on criteria B1ab(ii, iii, iv) and B2ab(ii, iii, iv). On the other hand, the relatively large EOO (1,700 km2) and the recent discovery of a new subpopulation implies that there might be some more undiscovered subpopulations within its potential range.